Constraint-based analysis¶
Simulation¶
framed implements several methods for simulating constraint-based models. Here are some examples:
from framed import FBA
solution = FBA(model)
print solution
Objective: 0.873921506968
Status: Optimal
All other simulation methods have a similar interface, just try:
from framed import pFBA, looplessFBA, MOMA, lMOMA, ROOM
If you have multiple solvers installed, you can tell framed to use a different solver:
from framed import set_default_solver
set_default_solver('cplex')
The simulation methods accept several additional arguments. Many of these arguments allow you to modify simulation parameters without changing the model.
For instance, you can easily change the model objective:
print FBA(model, objective={'R_ATPM': 1})
Objective: 175.0
Status: Optimal
You can also define additional constraints that will override the constraints in the model.
Note: The constraints can be specified as an interval or as a fixed value:
solution = pFBA(model, constraints={'R_EX_glc_e': (-5, 0), 'R_EX_o2_e': 0})
The solution object is interactive and you can use it for analysing different aspects of your results:
print solution.show_values(pattern='R_EX_')
R_EX_co2_e 22.8098
R_EX_glc_e -10
R_EX_h_e 17.5309
R_EX_h2o_e 29.1758
R_EX_nh4_e -4.76532
R_EX_o2_e -21.7995
R_EX_pi_e -3.2149
print solution.show_metabolite_balance('M_g6p_c', model)
[ --> o ] R_GLCpts 10
[ o --> ] R_Biomass_Ecoli_core_w_GAM -0.179154
[ o --> ] R_G6PDH2r -4.95998
[ o --> ] R_PGI -4.86086
print solution.show_metabolite_balance('M_g6p_c', model, percentage=True, sort=True)
[ --> o ] R_GLCpts 100.00%
[ o --> ] R_G6PDH2r -49.60%
[ o --> ] R_PGI -48.61%
[ o --> ] R_Biomass_Ecoli_core_w_GAM -1.79%
Flux variability analysis¶
To run flux variability analysis (FVA) just try:
from framed import FVA
result = FVA(model)
You can specify additional arguments such as required fraction of objective function
FVA(model, obj_percentage=0.9)
Since FVA is sensitive to the environmental conditions, you can easily override the model constraints:
FVA(model, constraints={'R_EX_o2': 0})
To speed-up computations, you can additionally specify a list of reactions that you are interested in analysing:
FVA(model, reactions=['R_PGI', 'R_PFK', 'R_PYK'])
If you just want to find and remove blocked reactions, then simply run:
from framed import blocked_reactions
blocked = blocked_reactions(model)
model.remove_reactions(blocked)
Better yet, if you can use the simplify method, which will also remove dead-end metabolites:
from framed import simplify
simplify(model)
Gene/reaction deletions¶
You can easily simulate gene and/or reaction deletions:
from framed import gene_deletion
genes = ['G_b2465', 'G_b2935']
solution = gene_deletion(model, genes)
from framed import reaction_deletion
reactions = ['R_PGI', 'R_PFK']
solution = reaction_deletion(model, reactions)
You can easily change the simulation method to any method available in framed:
gene_deletion(model, genes, method='pFBA')
gene_deletion(model, genes, method='MOMA')
gene_deletion(model, genes, method='lMOMA')
gene_deletion(model, genes, method='ROOM')
As always, you can easily override the model constraints without changing the model:
gene_deletion(model, genes, constraints={'R_EX_o2_e': 0})
Gene/reaction essentiality¶
You can calculate the set of essential genes (or reactions) as follows:
from framed import essential_genes
essential = essential_genes(model)
from framed import essential_reactions
essential = essential_reactions(model)
You can change the minimum growth threshold for which you consider a deletion to be lethal.
Let’s be more conservative and set it to 20% of the original growth rate:
essential_genes(model, min_growth=0.2)
Remember, gene (and reaction) essentiality is very much depedent on the environmental conditions.
Let’s try changing the carbon source:
essential_genes(model, constraints={'R_EX_glc_e': 0, 'R_EX_fru_e': (-10, 0)})
Omics-based methods¶
There are currently two simulation methods based on transcriptomics data implemented:
GIMME
from framed import GIMME
solution = GIMME(model, gene_expression)
Additional arguments (cutoff percentile, growth fraction) can be specified:
GIMME(model, gene_expression, cutoff=25, growth_frac=0.9)
E-Flux
from framed import eFlux:
solution = eFlux(model, gene_expression)
Additional arguments can be specified to scale the reaction rates to flux units using a measured reaction rate:
eFlux(model, gene_expression, scale_rxn='R_EX_glc_e', scale_value=11.5)
Strain design¶
framed doesn’t aim to be a strain design package. For that try Cameo instead.
Nonetheless, a few naive strain design methods are provided.
The first is a brute force approach that tries all possible combinations of gene/reaction deletions.
from framed import combinatorial_gene_deletion
objective = lambda x: x['R_EX_succ_e']
solutions = combinatorial_gene_deletion(model, objective, max_dels=3)
from framed import combinatorial_reaction_deletion
objective = lambda v: v['R_EX_succ_e']
solutions = combinatorial_reaction_deletion(model, objective, max_dels=3)
You can define more complex objective functions such as the BPCY, and you can easily change the simulation method:
biomass = model.detect_biomass_reaction()
BPCY = lambda v: v[biomass] * v['R_EX_succ_e'] / v['R_EX_glc_e']
solutions = combinatorial_gene_deletion(model, BPCY, method='MOMA', max_dels=3)
The other method is a heuristic hill-climbing approach:
from framed import greedy_gene_deletion
objective = lambda x: x['R_EX_succ_e']
solutions = greedy_gene_deletion(model, objective, max_dels=5)
from framed import greedy_reaction_deletion
objective = lambda x: x['R_EX_succ_e']
solutions = greedy_reaction_deletion(model, objective, max_dels=5)
You can fine-tune the balance between size of search space and speed by adjusting the population size:
solutions = greedy_gene_deletion(model, objective, max_dels=5, pop_size=20)
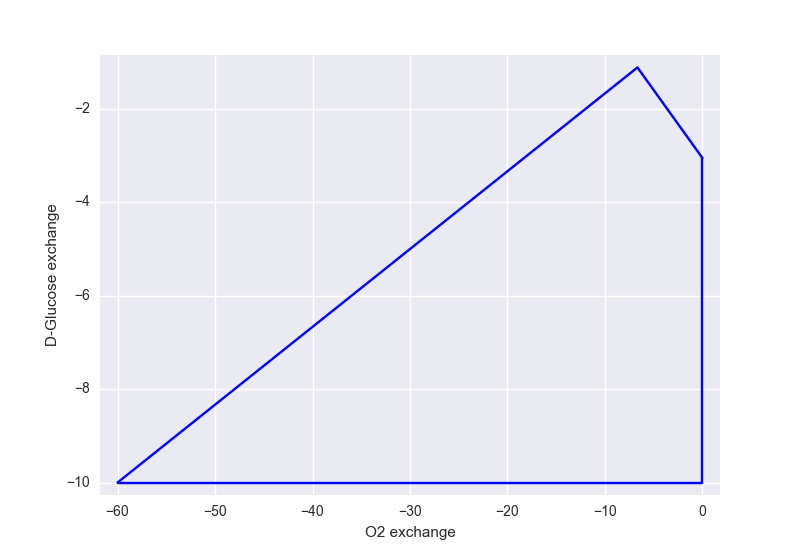
Plotting¶
framed provides some built-in plotting utilities. For instance, you can plot a flux envelope:
from framed import plot_flux_envelope
plot_flux_envelope(model, 'R_EX_o2_e', 'R_EX_glc_e')