Kinetic modeling¶
framed implements some basic (and experimental) support for working with kinetic models.
It now also supports models that contain assignment rules (see for example the Chassagnole 2002 E. coli model).
Simulation¶
Time-course¶
Running a simple time-course simulation (uses scipy odeint):
from framed import time_course
T, X = time_course(model, time=100)
You can change the number of integration steps:
time_course(model, time=100, steps=1000)
You can override model parameters without changing the model:
time_course(model, time=100, parameters={'Vmax1': 10.0, 'Vmax2': 20.0})
Steady-state¶
Find the steady-state metabolite concentrations and reaction rates:
from framed import find_steady_state
x_ss, v_ss = find_steady_state(model)
Again, you can easily override model parameters for simulation purposes:
find_steady_state(model, parameters={'Vmax1': 10.0})
Sampling¶
You can sample the steady-state solution space by manipulating the model parameters:
from framed import sample_kinetic_model
sample_kinetic_model(model, size=100)
You can define which parameters to sample:
sample_kinetic_model(model, size=100, parameters=['Vmax1', 'Vmax2'])
And you can define properties of the sampling distribution:
sample_kinetic_model(model, 100, distribution='normal', dist_args=(0, 1), log_scale=True)
Calibration¶
framed has basic support for calibrating kinetic models using time-course metabolomics data:
from framed import fit_from_metabolomics
fit_from_metabolomics(model, t_steps, data)
where data is a dictionary with time-course values for every measured metabolite.
You can specify which parameters you want to calibrate (by default it attempts to fit all parameters):
fit_from_metabolomics(model, t_steps, data, parameters=['Vmax1', 'Km1'])
and you can specify admissible bounds for the parameters:
fit_from_metabolomics(model, t_steps, data, bounds={'Vmax1': (0, 10), 'Km1': (0.1, 100)})
framed uses scipy’s minimize, function which implements several optimization methods. You can select the optimization method (see the scipy’s documentation for a list of available methods):
fit_from_metabolomics(model, t_steps, data, method='Nelder-Mead')
Plotting¶
framed implements some basic plotting utilities. For instance, you can plot a time-course simulation:
from framed import plot_timecourse
plot_timecourse(model, time=100)
You can define which metabolites you want to plot
plot_timecourse(model, time=100, metabolites=['S', 'P'])
You can overlay metabolomics data over your plots (for instance, after calibration):
plot_timecourse(model, time=100, data=my_data, data_steps=my_time_points)
As usual, you can override model parameters for a particular simulation without changing the model:
plot_timecourse(model, time=100, parameters={'Vmax': 10, 'Km': 0.1})
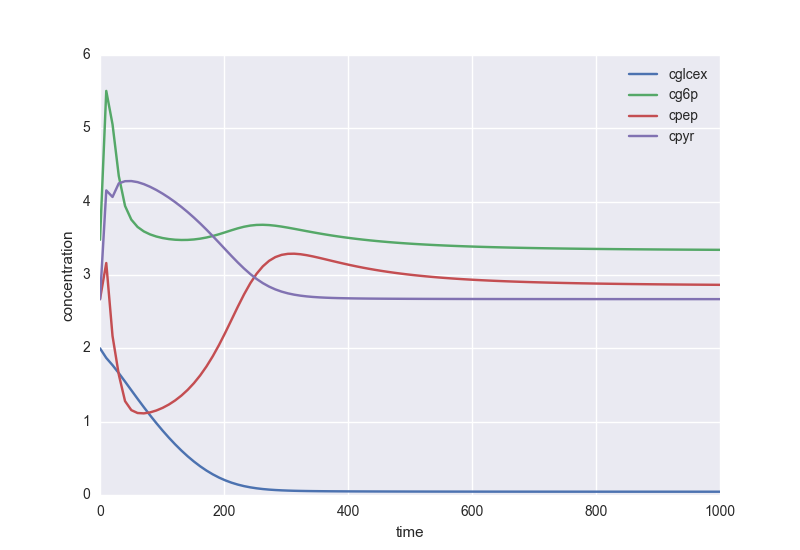
Here is a pratical example using the Chassagnole 2002 E. coli model:
plot_timecourse(model, 1e3, metabolites=['cglcex', 'cg6p', 'cpep', 'cpyr'],
xlabel='time', ylabel='concentration', parameters={'Dil': 0.2/3600})
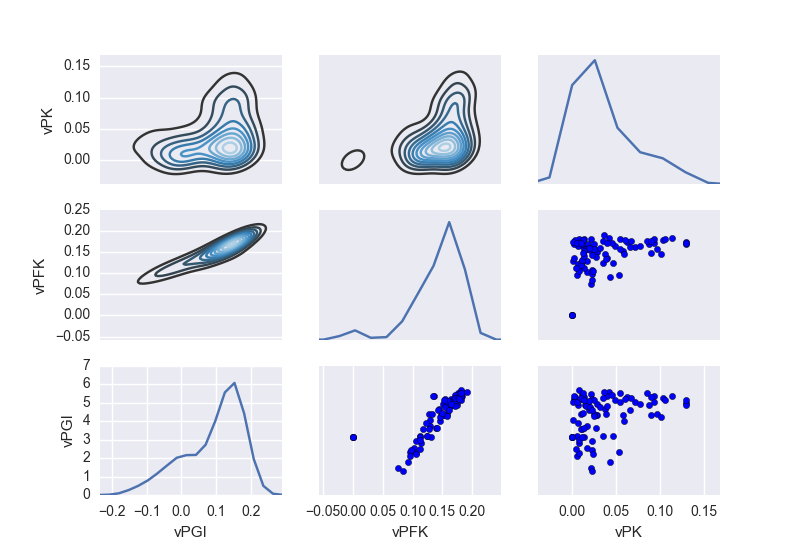
Finally, you can plot flux sampling results for a chosen set of reactions.
Note: this can also be applied to flux sampling results obtained with constraint-based models.
from framed import plot_flux_sampling
plot_flux_sampling(model, sample, reactions=['vPGI', 'vPFK', 'vPK'])